| 6.1 |
Class: Rel homology region factors (RHR) |
| Description: |
TRANSFAC® class description C0020: The structure of the Rel-type DBD exhibits a bipartite subdomain structure, each subdomain comprising a beta-barrel with five loops that form an extensive contact surface to the major groove of the DNA. Particularly, the first loop of the N-terminal subdomain (the highly conserved recognition loop) performs contacts with the recognition element on the DNA, but other loops are involved. The fact that the main DNA-contacts are made through loops has been suggested to provide a high degree of flexibility in binding to a range of different target sequences. Augmenting interactions are achieved by two alpha-helices within the N-terminal part that form strong minor groove contacts to the A/T-rich center of the B-element. In p65, the sequence between both alpha-helices is much shorter and even helix 2 is truncated. The second, C-terminal domain is necessary mainly for protein dimerization. |
| Aligned domain sequences: |
Alignment of RHR domains
Download fasta file Family 6.1.1: NF-kappaB-related factors
Download fasta file Family 6.1.3: NFAT-related factors
Download fasta file Family 6.1.4: CSL-related factors
Download fasta file Family 6.1.5: Early B-Cell Factor-related factors |
| Logo plot: |
Note: The four families of this class do not share significant sequence similarities. Therefore, logo plots of the DNA-binding domains of three of them are given for the four families distinctly. Family 6.1.4 (CSL-related factors) comprises only two sequences, therefore no logo plot is given for this clade.
Blue: basic residues (KRH); red: acidic residues (DE); purple: amides (QN); black: hydrophobic residues (AVLIPWFM); green: polar (CSTYG) (http://weblogo.berkeley.edu/) |
| Family 6.1.1: NF-kappaB-related factors |
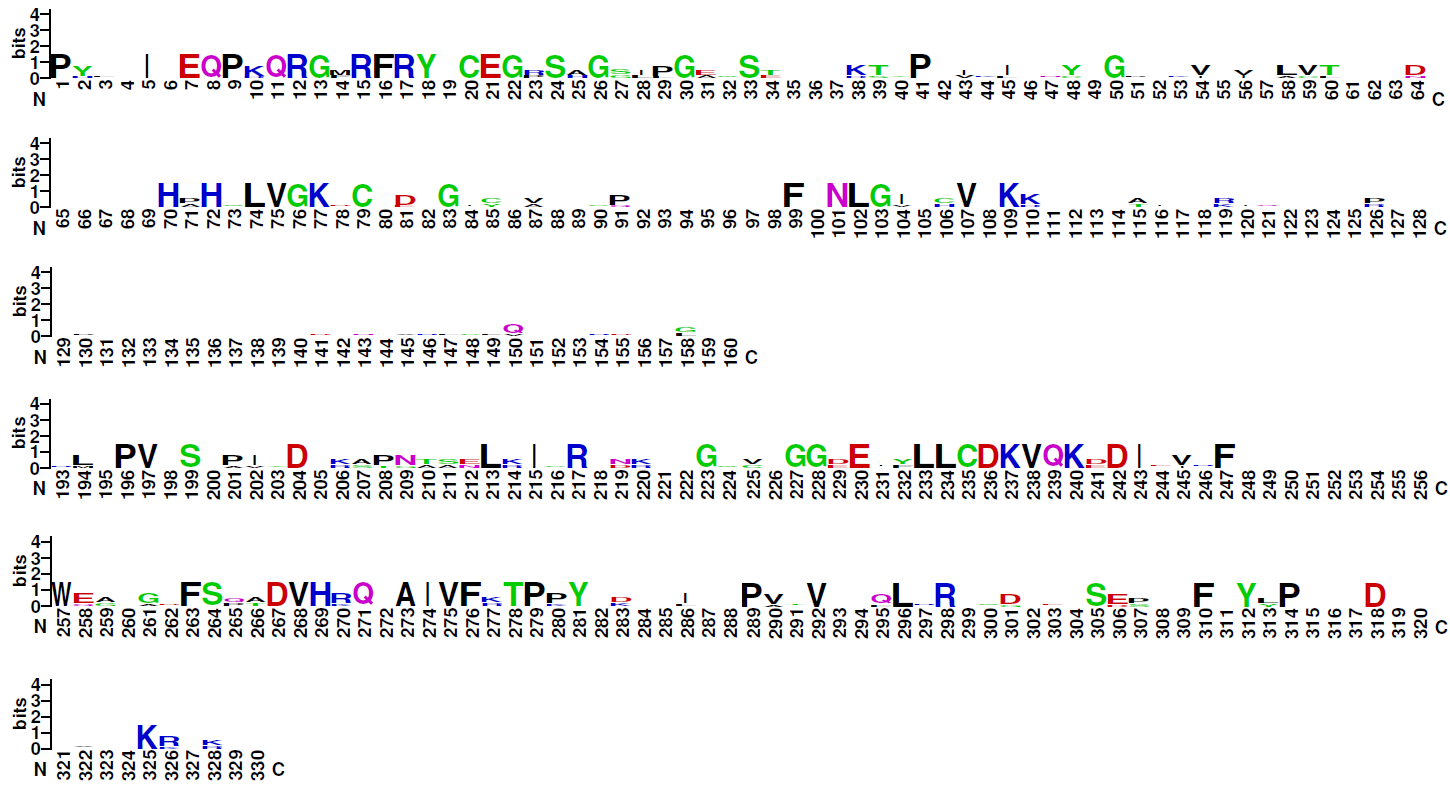
(The gap between positions 160 and 193, which have been skipped from the logo shown, is due to long insertions in most of the aligned sequences.) |
| Family 6.1.3: NFAT-related factors |
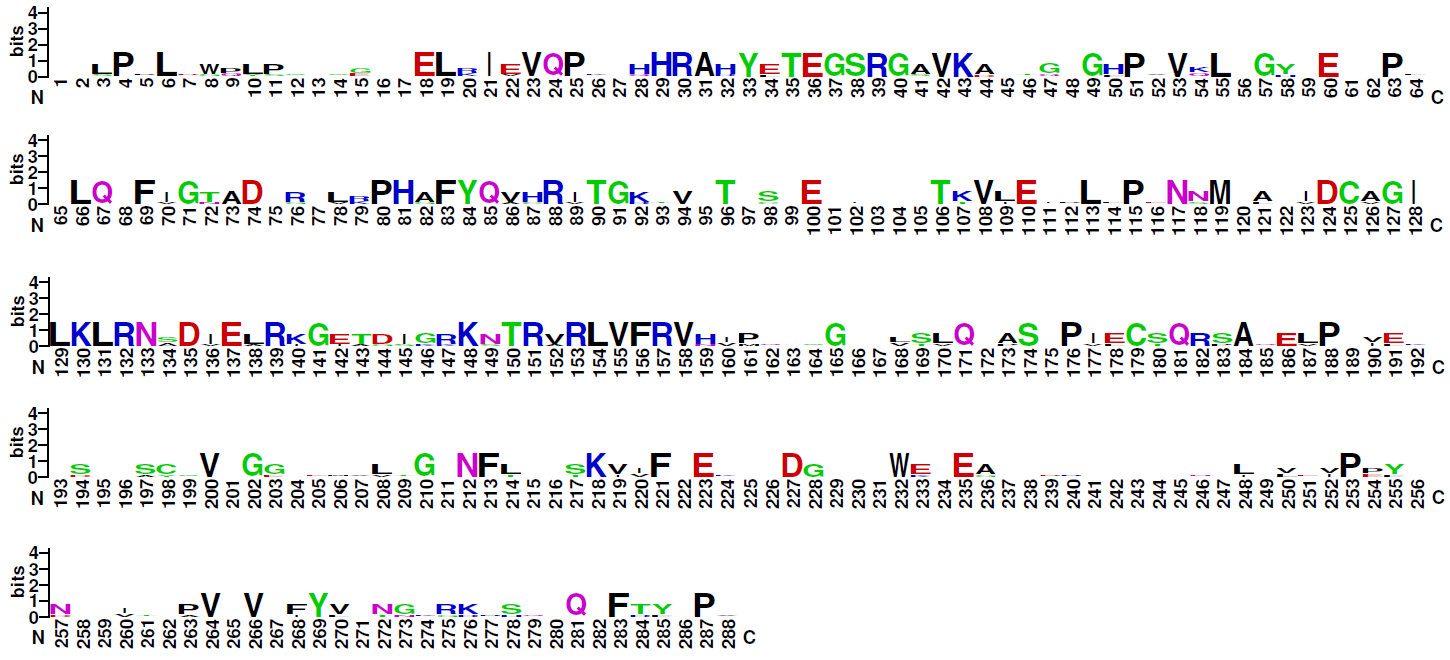 |
| Family 6.1.5: Early B-Cell Factor-related factors |
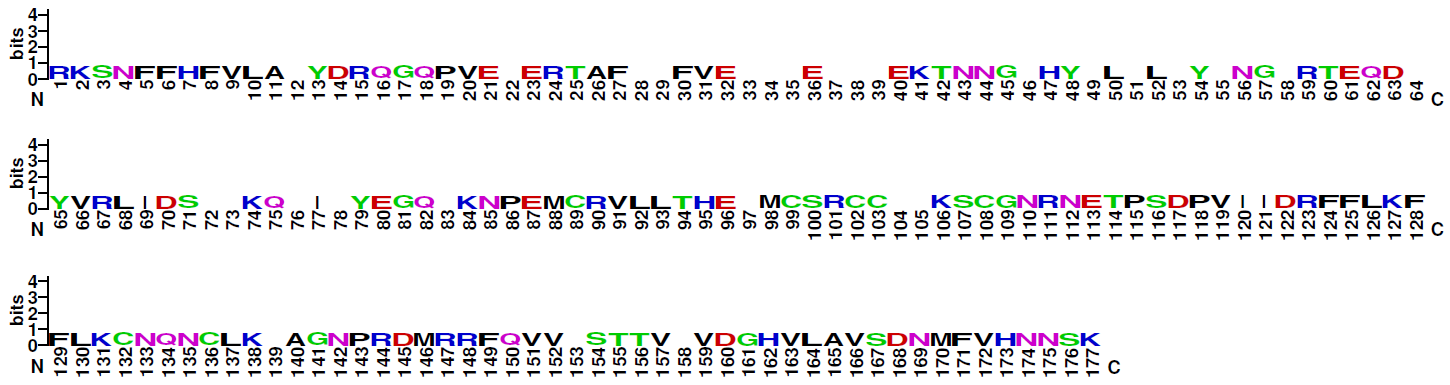 |


